Project Overview
|
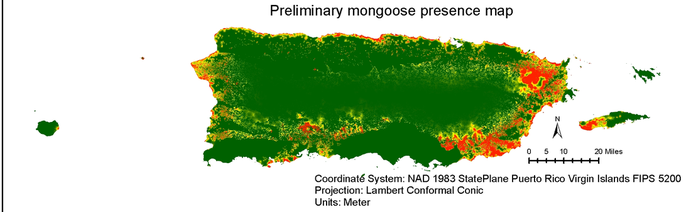
The overall objective of my project is to understand the influence of ecological processes on the genetic variation of the small Asian mongoose (Herpestes auropunctatus) in Puerto Rico and the US Virgin Islands, and explore dietary preferences of mongooses in important nesting habitats for nesting birds and turtles.
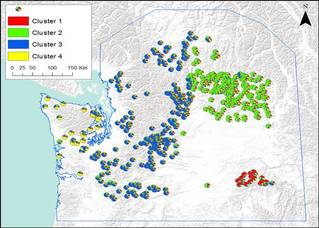
I will quantify the relationship between landscape variables and population genetic structure to ultimately understand how landscape features and adaptive processes such as gene flow, genetic drift and selection drive the degree and the spatial distribution of mongoose genetic variation. Using a landscape genetics I can use spatially-explicit techniques will allow me to identify landscape barriers which can disrupt gene flow, that wouldn't be detected otherwise by traditional population genetic methods. |
Landscape genetic studies require data from two sources: landscape data (i.e. remotely sensed data) and multi-locus genetic data. For the small Asian mongoose, 9 polymorphic microsatellite loci have been developed in 2009 by Thulin et al. , which were true to the HW equilibrium.
On the diet...
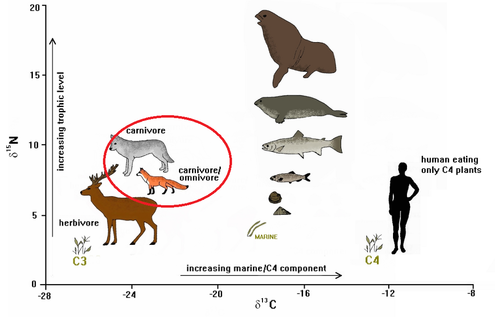
I will examine feeding preferences of mongooses in important coastal nesting sites of three species of turtle (leatherback, green and hawksbill) as a function of location from coastal forest edge to interior and season, identify the trophic position of the mongoose in this system, and locate critical areas where mongoose diet is substantially composed of turtle hatchlings.
Understanding how mongooses partition their resources and identifying when and where turtle nests are most prone to predation, conservation managers can increase the chances of turtle hatchling survival by targeting high predation risk areas. Additionally, a better understanding of mongoose trophic links allows for inferences on the vulnerability of invaded vertebrate communities.
Understanding how mongooses partition their resources and identifying when and where turtle nests are most prone to predation, conservation managers can increase the chances of turtle hatchling survival by targeting high predation risk areas. Additionally, a better understanding of mongoose trophic links allows for inferences on the vulnerability of invaded vertebrate communities.